The interaction of ribonucleic acid and water
MBI scientists use vibrational spectroscopy to analyze biomolecular systems
Ribonucleic acid (RNA) plays a key role in biochemical processes which occur at the cellular level in a water environment. Mechanisms and dynamics of the interaction between RNA and water were now revealed by vibrational spectroscopy on ultrashort time scales and analyzed by in-depth theory
Ribonucleic acid (RNA) represents an elementary constituent of biological cells. While deoxyribonucleic acid (DNA) serves as the carrier of genetic information, RNA displays a much more complex biochemical functionality. This includes the transmission of information in the form of mRNA, RNA-mediated catalytic function in ribosomes, and the encoding of genetic information in viruses. RNA consists of a sequence of organic nucleobase molecules which are held together by a so-called backbone consisting of phosphate and sugar groups.
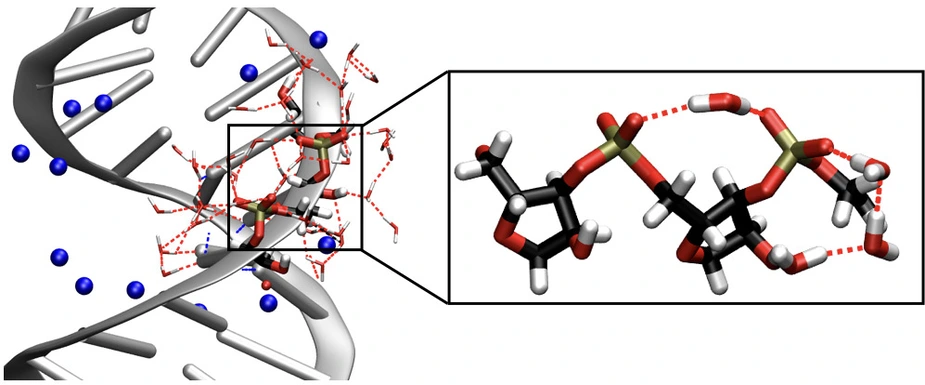
Such a sequence can exist as a single strand or in a paired double-helix geometry. Both forms are embedded in a water shell and their phosphate and sugar groups are distinct docking points for water molecules. The structure of the water shell fluctuates on a time scale of a few tenth of a picosecond (1 ps = 10-12 s = 1 millionth of a millionth of a second). The interactions of RNA and water and their role for the formation of three-dimensional RNA structures are only understood insufficiently and difficult to access by experiment.
Scientists from the Max Born Institute have now observed the interaction of RNA with its water shell in real time. In their new experimental method, vibrations of the RNA backbone serve as sensitive noninvasive probes of the influence of neighboring water molecules on the structure and dynamics of RNA. The so-called two-dimensional infrared spectroscopy allows for mapping the time evolution of vibrational excitations and for determining molecular interactions within RNA and between RNA and water. The results show that water molecules at the RNA surface perform tipping motions, so-called librations, within a fraction of a picosecond whereas their local spatial arrangement is preserved for a time range longer than 10 ps. This behavior deviates strongly from that of neat water and is governed by the steric boundary conditions set by the RNA surface. Individual water molecules connect neighboring phosphate groups and form a partly ordered structure which is mediated by their coupling to the sugar units.
The librating water molecules generate an electrical force by which the water fluctuations are transferred to the vibrations of RNA. The different backbone vibrations display a diverse dynamical behavior which is determined by their local water environment and reflects its heterogeneity. RNA vibrations also couple mutually and exchange energy among themselves and with the water shell. The resulting ultrafast redistribution of excess energy is essential for avoiding a local overheating of the sensitive macromolecular structure. This complex scenario was analyzed by detailed theoretical calculations and simulations which, among other results, allowed for the first complete and quantitative identification of the different vibrations of the RNA backbone. Comparative experiments with DNA reveal similarities and characteristic differences between these two elementary biomolecules, showing a more structured water arrangement around RNA.
The study highlights the strong potential of non-invasive time-resolved vibrational spectroscopy for unraveling the interplay of structure and dynamics in complex biomolecular systems on molecular length and time scales.
Original Publication:
E. M. Bruening, J. Schauss, T. Siebert, B. P. Fingerhut, T. Elsaesser: Vibrational Dynamics and Couplings of the Hydrated RNA Backbone: A Two-Dimensional Infrared Study.
J. Phys. Chem. Lett. 9, 583-587 (2018). DOI: 10.1021/acs.jpclett.7b03314.
Contact:
Max-Born-Institut für Nichtlineare Optik und Kurzzeitspektroskopie (MBI)
Forschungsverbund Berlin e.V.
Dr. Benjamin Fingerhut
Tel.: 030 6392-1404
fingerhut(at)mbi-berlin.de
Prof. Dr. Thomas Elsaesser
Tel.: 030 6392-1400
elsasser(at)mbi-berlin.de