Coronavirus: BESSY II data accelerate drug development
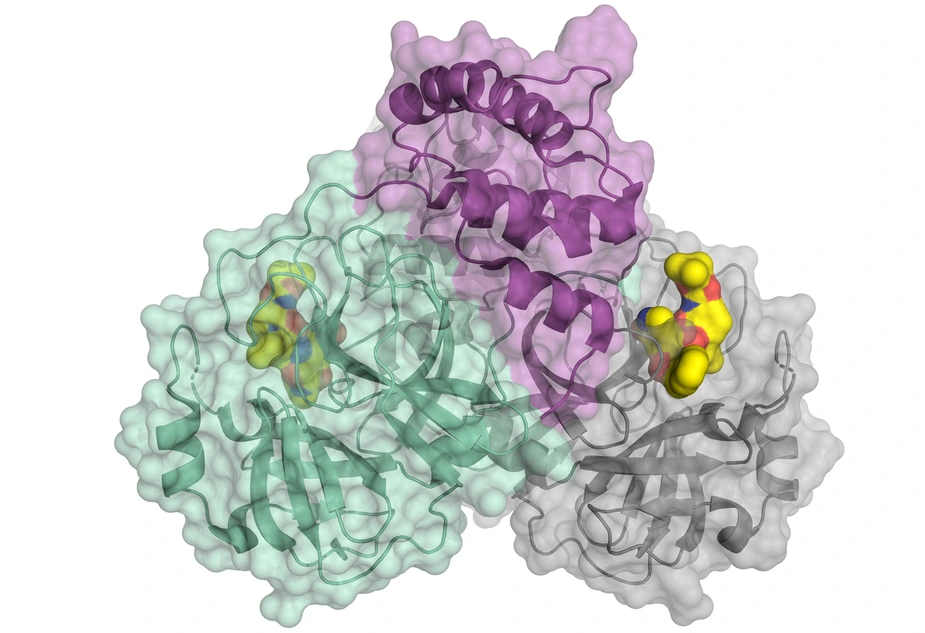
Scientists have decoded the three-dimensional architecture of the main protease of SARS-CoV-2
A coronavirus is keeping the world in suspense. SARS-CoV-2 is highly infectious and can cause severe pneumonia with respiratory distress (COVID-19). Scientists are doing research in order to prevent the viruses from multiplying. A team from the University of Lübeck has now found a promising approach. Using the high-intensity X-ray light from the Berlin synchrotron source BESSY II, they have decoded the three-dimensional architecture of the main protease of SARS-CoV-2. This protein plays a central role in the reproduction of the virus. Analysing the 3D architecture allows the systematic development of drugs which inhibit the reproduction oft he virus.
Teams around the world are working hard to develop active substances against SARS-CoV-2. The structural analysis of functional proteins of the virus is very helpful for this goal. The function of a protein is closely related to its 3D architecture. If this 3D architecture is known, it is possible to identify specific points of attack for active substances.
Inhibiting the reproduction
A special protein is responsible for the reproduction of the viruses: the viral main protease (Mpro or also 3CLpro). A team led by Prof. Dr. Rolf Hilgenfeld, University of Lübeck, has now decoded the 3D architecture of the main protease of SARS-CoV-2. The researchers have used the high-intensity X-ray light from the BESSY II facility of the Helmholtz-Zentrum Berlin.
Fast track at BESSY II
"For such issues of highest relevance, we provide fast track access to our instruments", says Dr. Manfred Weiss, who heads the Research Group Macromolecular Crystallography (MX) at HZB. At the so-called MX instruments tiny protein crystals can be analysed with highly brilliant X-ray light. The images contain information about the 3D architecture of the protein molecules. The complex shape of the protein molecule and its electron density is then calculated by computer algorithms.
Targets for active substances
The 3D architecture provides concrete starting points for developing active substances or inhibitors. These drugs could dock specifically to target points of the macromolecule and impede its function. Rolf Hilgenfeld is a world-renowned expert in the field of virology and already developed an inhibitor against the SARS-virus during the 2002/2003 SARS pandemic. In 2016, he succeeded in deciphering an enzyme of the Zika virus.
The results are published in advance in a preprint version in biorxiv.org (without peer review) so that the data and results are available to all researchers worldwide. The study will be published (with peer review) on 20. March 2020 in Science.
"X-ray Structure of Main Protease of the Novel Coronavirus SARS-CoV-2 Enables Design of α-Ketoamide Inhibitors"; Linlin Zhang, Daizong Lin, Xinyuanyuan Sun, Katharina Rox, Rolf Hilgenfeld
DOI: 10.1101/2020.02.17.952879
For further information and a filmclip visit
www.helmholtz-berlin.de/pubbin/news_seite
Contact:
Helmholtz-Zentrum Berlin für Materialien und Energie
Dr. Manfred Weiss
Research Group Macromolecular Crystallography
Head of MX group at BESSY II
Phone: +49 30 8062-13149
Email: manfred.weiss(at)helmholtz-berlin.de
Press Officer:
Dr. Antonia Rötger
Phone: +49 30 8062-43733
Email: antonia.roetger(at)helmholtz-berlin.de